Data
Data: How to surf your data, rather than drown in it!
Introduction
Nuts & Bolts
This first section covers some basic, but perhaps surprising, aspects of working with files. How best to copy them or move them from one computer to another on the network. We'll also compare the properties of some different file formats.
Data on Disk
A Salutary Tale of Copying Files
We'll start by considering data stored on a disk drive. One thing that you might not know is that file systems and disk drives perform best when they are dealing with larger files. But how large is large? Here's a simple example, which you can try yourself:
First of all, let's get ourselves a large-ish file. A compressed tarball of the source code for the Linux kernel will do. In this case it's about 70MB in size. We can time how long it takes to create a copy of it. Below are the results of doing this experiment on BlueCrystal phase 2:
BCp2$ wget https://www.kernel.org/pub/linux/kernel/v3.x/testing/linux-3.10-rc7.tar.xz BCp2$ time cp linux-3.10-rc7.tar.xz linux-3.10-rc7.tar.xz.copy real 0m3.530s user 0m0.000s sys 0m0.068s
Now, that tar ball contains around 47,000 files, many of which are only a few hundred or thousand of bytes in size. These are files at the smaller end of the scale. Let's unpack the tarball and time how long it takes to copy these files, one-by-one:
BCp2$ tar --use-compress-program=xz -xf linux-3.10-rc7.tar.xz BCp2$ time cp -r linux-3.10-rc7 linux-3.10-rc7.copy real 18m17.102s user 0m0.214s sys 0m6.359s
Yikes! that took over 350 times longer than copying the single, large file. (These timings were taken at ~10:45 on the 25 Jun 2013. Any differences from the above values will be due to differences in load on the filesystem, which is shared with all the other users of the cluster. More of that in a moment..)
Now, we can repeat these tests on a different system. I got the values below from my very standard desktop machine:
desktop$ wget https://www.kernel.org/pub/linux/kernel/v3.x/testing/linux-3.10-rc7.tar.xz desktop$ time cp linux-3.10-rc7.tar.xz linux-3.10-rc7.tar.xz.copy real 0m0.192s user 0m0.000s sys 0m0.156s desktop$ tar --use-compress-program=xz -xf linux-3.10-rc7.tar.xz desktop$ time cp -r linux-3.10-rc7 linux-3.10-rc7.copy real 0m25.961s user 0m0.168s sys 0m2.360s
That's a lot quicker! However, copying the small files still took over 130 times longer than copying the large file. (Again, your mileage may vary.)
But hang on, I thought BlueCrystal was meant to be "super"?! Well it is. It's just that it's filesystem is servicing much more than just your file copying request.
Modern SATA disks have read and write bandwidths close to 100MB/s--for example, I just tested my Linux desktop machine with a handy, built-in utility (System > Administration > Disk utility) and recorded a read performance of ~75MB/s. We can compare this to the filesystem on BlueCrystal phase 2, where we see a peak of about 500MB/s throughput on the mixed workload of full, running cluster.
Another test below highlights a key difference between a parallel filesystem and that on a single disk. If I start several processes writing to the disk in my desktop machine, I see a rapid drop-off in performance as the number of processes increases. In contrast the parallel filesystem is able to support many processes with modest degradation to file writing performance.
| System | Clients x1 | Clients x4 | Clients x8 |
| Desktop | ~65MB/s | ~10MB/s | ~0.5MB/s |
| Parallel FS | ~160MB/s | 130MB/s | 67MB/s |
File Formats and File Sizes
The amount of disk space that a file consumes depends a great deal on it's format. As an example, I have created two files--one binary format and the other ASCII text format. Both files contain the same sequence of floating point numbers from 1.0 to 5000.0 (1.0, 2.0, 3.0 etc.). We can satisfy ourselves that they contain the same data by inspecting their contents. This is simple for the text file:
less output.txt 1.0000000 2.0000000 3.0000000 4.0000000 5.0000000 6.0000000 7.0000000 ...
but a little more involved for the binary file. For this I'm using the Linux octal dump (od) command, with the appropriate format flag:
od -f output.bin | less 0000000 2.802597e-41 1.000000e+00 2.000000e+00 3.000000e+00 0000020 4.000000e+00 5.000000e+00 6.000000e+00 7.000000e+00 ...
A long listing reveals the size difference for the two files:
-rw-r--r-- 1 gethin gethin 220016 2013-07-23 16:44 output.bin -rw-r--r-- 1 gethin gethin 880002 2013-07-23 16:44 output.txt
output.bin is around 220KB in size, whereas output.txt is around 4x that size! The text format is convenient to read, but is a lot less space efficient.
We can experiment with compressing the text file, in this case with the Linux gzip tool:
gzip output.txt
After compression, the size of the text file rivals that of the binary version:
-rw-r--r-- 1 gethin gethin 223780 2013-07-23 16:44 output.txt.gz
Using Local Disk on BlueCrystal Nodes
If you are using BlueCrystal, you may want to consider using disks local to the compute nodes. These nodes are 300GB in size and are accessible via /local. If you do elect to use local disks:
- It is advisable to check the available space in /local as part of your job.
- Please do clean up /local after your job has finished
- When transferring data from node to node, please use the suffix .data.cluster when referring to a node, e.g. scp u04n037.data.cluster:/local/mydata $HOME.
Data over the Network
If you are copying or moving data over the network, you will minimise the amount of time that you'll have to wait by making best use of the network bandwidth that is available. It may be surprising to learn that the size of files that you are working with will have a direct impact upon the network bandwidth that you can acquire.
An analogy between computer networks and pipelines and plumbing is a useful one, including notions of pipeline capacity, bottlenecks etc.
A pipe has a capacity to carry a certain volume of liquid per unit of time. It makes sense that a wider pipe has a greater carrying capacity. It is also intuitive that if we send a trickle of water down the pipe, we won't be exploiting it's full capacity.
Moving files from one computer to another typically involves reading from the hard disk drive on the source machine, transmitting the data down the network and depositing it onto the destination's disk. Since technology dictates that we read, write and transmit data a different speeds, we could think of this end-to-end arrangement as a sequence of different width pipes all connected together. That being the case, where is the bottleneck and how does that limit the rate at which we can move data around?
Let's examine some numbers. A common networking technology is gigabit ethernet (gigE). As it's name suggests, it can--in principle--transmit data at a rate of 1,000,000,000 bits per second, or 125MB/s. However, some details of the switching technology means that in practice transmission rates of around 35MB/s are seen. So is the switching the bottleneck in our system? Well, it depends.
Let's try copying a large file over a gigE network. the Linux scp command obligingly gives us a transmission rate for the transaction:
linux-3.10-rc7.tar.xz 100% 70MB 23.3MB/s 00:03
Aha. OK. So it's not quite up at 35MB/s, but it's in the right ballpark.
Let's repeat the experiment with the ruck of small files that is contained within the tar archive:
.gitignore 100% 1091 1.1KB/s 00:00 .mailmap 100% 4465 4.4KB/s 00:00 COPYING 100% 18KB 18.3KB/s 00:00 CREDITS 100% 93KB 93.1KB/s 00:00 BUG-HUNTING 100% 8326 8.1KB/s 00:00 CodingStyle 100% 31KB 30.7KB/s 00:00 DMA-API-HOWTO.txt 100% 31KB 31.3KB/s 00:01 DMA-API.txt 100% 27KB 27.3KB/s 00:00 ...
Ouch! that is some pretty terrible throughput. Just to reinforce the point, copying the large file took around 3 seconds, whereas I stopped the second experiment after 20 mins had elapsed.
So what's the story? In the second experiment, the bottleneck is the process of finding each individual file on the source disk and creating a copy on the destination disk. That being the case, in terms of our analogy, we're only able to trickle the fluid into the pipe and so the best case carrying capacity is missed by a large margin.
The lesson here is that you need to transmit large files to be able to exploit the full network bandwidth--and this will be even more the case if you move to faster, i.e. greater carrying capacity, networks such as 10gigE.
If you are stuck with a bunch of small files to transmit, you could potentially reap a reward by creating a tar file on-the-fly with a pipelined Linux command such as:
tar zcvf - /path/to/dir/of/small-files | ssh user@destination.computer "cat > /destination/dir/my-archive.tar.gz"
Analysing Data
OK, let's assume that you have all your data where you want it and in the most suitable file format. Now we can begin to actually work on the data. Let's take a moment to consider the kind of things you might want to do:
- Cleaning and filtering: Raw data may contain instrument noise, typos, missing values etc. That being the case, it's likely that we'll need to clean it up somewhat before use. The act of cleaning could include finding & correcting typos, inserting a missing data symbol, interpolating or even removing outliers. Linux can provide tools to help with many of these tasks. Regular expressions constitute a powerful language for pattern matching and tools such as sed and awk are two tools which can used to apply them.
- Sorting and searching: Computer scientists spend a lot of time thinking about the most efficient ways to sort data and to search collections for given values. Why? Well, these are relatively common operations to be done on a computer. Think of web searches, cinema listings, price ordered shopping options etc. Databases (introduced in the next section) are particularly useful and efficient tools for these kinds of operations. Sorting and searching go together as it can be much quicker to search through a sorted list of items, rather than one in random order. More on this and other concepts, such as hash tables in the section on writing your own code.
- Calculating summary statics: Once we've selected the items of interest, we might wish to summarise their characteristics. For example calculate means, medians and variances. Their are many good tools to help you do this, including the languages/packages R, Python and Matlab.
- Creating plots & graphics: A picture can be worth a thousand words. Again R, Python and Matlab can help here.
- Tests of statistical significance: Perhaps you've collected data from two populations--one group, who are receiving a new drug and the other, who are given a placebo. Can you prove that your observations of the two groups really are different, and not just the result of random variation? You've guessed it, R, Python and Matlab can help!
Databases
It may be tempting to store your data in many small (text) files. Perhaps this provides an intuitive way to catalogue the information. However, we saw above that you will suffer a performance penalty if you store your data this way. If your data is amenable to being stored in tabular form, i.e. in rows and columns, then you may well be better off using one of many available database packages. These packages are optimised for search performance and excel at random access patterns for both reading and writing data.
SQL (the Structured Query Language) is a language designed for working with databases and is common to the examples given below.
SQLite
A very popular relational database. Lightweight as it is unmanaged and so has very simple access controls. However does support SQL. The command line interface is widely available. For example, it is installed on BlueCrystal.
Let's start up the command line interface:
sqlite3 test.db
In this case, we've specified the file test.db. If the file exists, it will open it. Else it will create a new database to be stored in the given file.
Without further ado, let's create a table in given database and populate it with some records:
sqlite> CREATE TABLE planets(Id INT, Name TEXT, Diameter REAL, Mass REAL, Orbital_Period REAL);
sqlite> INSERT INTO planets VALUES(1,'Mercury',0.382,0.06,0.24);
sqlite> INSERT INTO planets VALUES(2,'Venus',0.949,0.82,0.72);
sqlite> INSERT INTO planets VALUES(3,'Earth',1.0,1.0,1.0);
sqlite> INSERT INTO planets VALUES(4,'Mars',0.532,0.11,1.52);
sqlite> INSERT INTO planets VALUES(5,'Jupiter',11.209,317.8,5.20);
sqlite> INSERT INTO planets VALUES(6,'Saturn',9.449,95.2,9.54);
sqlite> INSERT INTO planets VALUES(7,'Uranus',4.007,14.6,19.22);
sqlite> INSERT INTO planets VALUES(8,'Neptune',3.883,17.2,30.06);Now, let's see the fruits of our labours. After setting some formatting information, we can use an SQL command to select all the records in the planets table:
sqlite> .mode column
sqlite> .headers on
sqlite> SELECT * FROM planets;Id Name Diameter Mass Orbital_Period ---------- ---------- ---------- ---------- -------------- 1 Mercury 0.382 0.06 0.24 2 Venus 0.949 0.82 0.72 3 Earth 1.0 1.0 1.0 4 Mars 0.532 0.11 1.52 5 Jupiter 11.209 317.8 5.2 6 Saturn 9.449 95.2 9.54 7 Uranus 4.007 14.6 19.22 8 Neptune 3.883 17.2 30.06
We can also issue a more exacting query. In this case, let's ask for all the planets which have a mass greater than or equal to that of the Earth:
sqlite> SELECT * FROM planets WHERE Mass >= 1.0;Id Name Diameter Mass Orbital_Period ---------- ---------- ---------- ---------- -------------- 3 Earth 1.0 1.0 1.0 5 Jupiter 11.209 317.8 5.2 6 Saturn 9.449 95.2 9.54 7 Uranus 4.007 14.6 19.22 8 Neptune 3.883 17.2 30.06
So far so good. Let's create an additional table called moons:
sqlite> CREATE TABLE moons(Name TEXT, Num_Moons INT);
sqlite> INSERT INTO moons VALUES('Mercury',0);
sqlite> INSERT INTO moons VALUES('Venus',0);
sqlite> INSERT INTO moons VALUES('Earth',1);
sqlite> INSERT INTO moons VALUES('Mars',2);
sqlite> INSERT INTO moons VALUES('Jupiter',67);
sqlite> INSERT INTO moons VALUES('Saturn',62);
sqlite> INSERT INTO moons VALUES('Uranus',27);
sqlite> INSERT INTO moons VALUES('Neptune',13);
sqlite> INSERT INTO moons VALUES('Pluto',5);Now that we have two tables, we can examine the very powerful feature of JOINing tables , common to all good relational databases. A, so called, natural inner join will create a new table, on-the-fly, from all the records in the joined tables which have matching values:
sqlite> SELECT Name, Orbital_Period, Num_moons FROM planets NATURAL JOIN moons;Name Orbital_Period Num_Moons ---------- -------------- ---------- Mercury 0.24 0 Venus 0.72 0 Earth 1.0 1 Mars 1.52 2 Jupiter 5.2 67 Saturn 9.54 62 Uranus 19.22 27 Neptune 30.06 13
Note that Pluto was not listed as a result of the inner join, since it is not present in the planets table.
We can also create an outer join, which is not so constrained. SQLite does not, as yet, support a 'right outer join' and so I needed to swap the order of the tables in the join so that my 'left outer join' contained all those in the left table (i.e. the table containing the name Pluto).
sqlite> SELECT Name, Orbital_Period, Num_moons FROM moons NATURAL LEFT OUTER JOIN planets;Notice that Pluto record is blank in the Orbital_Period column as there is no corresponding value in the planets table.
Name Orbital_Period Num_Moons ---------- -------------- ---------- Mercury 0.24 0 Venus 0.72 0 Earth 1.0 1 Mars 1.52 2 Jupiter 5.2 67 Saturn 9.54 62 Uranus 19.22 27 Neptune 30.06 13 Pluto 5
If you would like to export to, e.g., a CSV file (this is useful for subsequent import into, e.g., R):
sqlite> .mode csv
sqlite> .output planets.csv
sqlite> SELECT * FROM planets WHERE Mass >= 1.0;Where the contents of the file planets.csv is:
Id,Name,Diameter,Mass,Orbital 3,Earth,1.0,1.0,1.0 5,Jupiter,11.209,317.8,5.2 6,Saturn,9.449,95.2,9.54 7,Uranus,4.007,14.6,19.22 8,Neptune,3.883,17.2,30.06
There is, of course a corresponding command to import data from a file into a table. For example, if I had information about stars in an appropriately formatted CSV files, I could load it into a table called stars using the commands:
sqlite> .separator ','
sqlite> .import stars.csv starsTo exit the SQLite command line interpreter type:
sqlite> .exitIt is perfectly possible--indeed, perhaps preferable--to access a database from inside a program. (SQLite was really designed with that in mind.) For example, you can learn more about accessing an SQLite database from inside a python script at:
More information about SQLite and other interfaces for access is at:
- SQLite: e.g. http://zetcode.com/db/sqlite/
MySQL
Taking a step up on the functionality ladder, MySQL is a popular, open-source, enterprise grade relational database management system (RDBMS), which is readily available for most operating systems. In the notes below, I will assume that you have MySQL installed and have set a password for the root user. For popular Linux distributions, such as Ubuntu and CentOS, MySQL is easily installed through the package manager. (More information on MySQL installation.)
OK, our first tasks will be to connect to the MySQL monitor tool as the administrator, and to create new users and databases:
gethin@gethin-desktop:~$ mysql -u root -p Enter password:
After typing the administrator password we are greeted with:
Welcome to the MySQL monitor. Commands end with ; or \g. Your MySQL connection id is 36 Server version: 5.1.69-0ubuntu0.10.04.1 (Ubuntu) Copyright (c) 2000, 2013, Oracle and/or its affiliates. All rights reserved. Oracle is a registered trademark of Oracle Corporation and/or its affiliates. Other names may be trademarks of their respective owners. Type 'help;' or '\h' for help. Type '\c' to clear the current input statement. mysql>
Let's create a user. Note that the password, changeme in this case, is in clear text.
mysql> CREATE USER 'gethin'@'localhost' IDENTIFIED BY 'changeme';
Query OK, 0 rows affected (0.03 sec)A database, called menagerie, to practice with:
mysql> CREATE DATABASE menagerie;
Query OK, 1 row affected (0.00 sec)And we'll grant some privileges (in this case ALL--a full set of privileges) for all the tables to be stored in the menagerie database to the user called gethin:
mysql> GRANT ALL ON menagerie.* TO 'gethin'@'localhost';
Query OK, 0 rows affected (0.02 sec)Now, to work on that database, we want to disconnect as the administrator and connect as an appropriate user:
gethin@gethin-desktop:~$ mysql -u gethin -p
Enter password:switch to the database in question:
mysql> use menagerie
Database changedand create a new table:
mysql> CREATE TABLE planets (Id INT NOT NULL AUTO_INCREMENT,
-> Name VARCHAR(10),
-> Diameter REAL,
-> Mass REAL,
-> Orbital_Period REAL,
-> PRIMARY KEY (Id)
-> );
Query OK, 0 rows affected (0.00 sec)Now, if we had a CSV file of the form:
1,'Mercury',0.382,0.06,0.24 2,'Venus',0.949,0.82,0.72 3,'Earth',1.0,1.0,1.0 4,'Mars',0.532,0.11,1.52 5,'Jupiter',11.209,317.8,5.20 6,'Saturn',9.449,95.2,9.54 7,'Uranus',4.007,14.6,19.22 8,'Neptune',3.883,17.2,30.06
we could load this directly into our planets table, without the labourious SQL INSERT commands:
LOAD DATA LOCAL INFILE 'planets.csv' INTO TABLE planets FIELDS TERMINATED BY ',';et voila, we can see the data duly loaded into the table:
mysql> SELECT * FROM planets;
+----+-----------+----------+-------+----------------+
| Id | Name | Diameter | Mass | Orbital_Period |
+----+-----------+----------+-------+----------------+
| 1 | 'Mercury' | 0.382 | 0.06 | 0.24 |
| 2 | 'Venus' | 0.949 | 0.82 | 0.72 |
| 3 | 'Earth' | 1 | 1 | 1 |
| 4 | 'Mars' | 0.532 | 0.11 | 1.52 |
| 5 | 'Jupiter' | 11.209 | 317.8 | 5.2 |
| 6 | 'Saturn' | 9.449 | 95.2 | 9.54 |
| 7 | 'Uranus' | 4.007 | 14.6 | 19.22 |
| 8 | 'Neptune' | 3.883 | 17.2 | 30.06 |
+----+-----------+----------+-------+----------------+
8 rows in set (0.00 sec)Writing Efficient Queries
This is an important area that I would like to develop. If you have a case study, I'd be interested to hear from you so that I can develop some relevant notes on this topic.
UoB Data Haven
The University maintains a central Oracle database which is available to members of staff to store their data in a managed and backed-up environment. To learn more, visit: http://www.bris.ac.uk/it-services/applications/infosystems/datahaven/.
GUI Tools
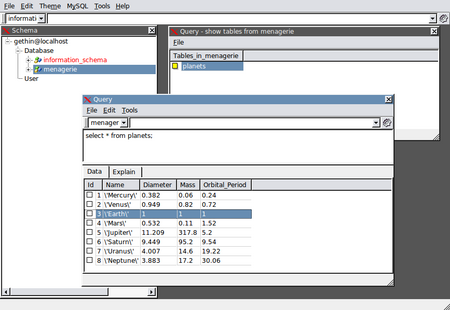
If you would prefer to access your data and tables via a GUI, SQL Navigator is a popular, cross-platform tool. The image below shows access to the planets table in the menagerie database:
The University also runs courses on learning and using MS Access. More information visit: http://www.bris.ac.uk/it-services/learning/staff.html.
Rolling Your Own
OK, so you've perused the available tools, programs and packages and you've decided that the best plan is for you to write your own code--good on ya! Now, when thinking about processing data, there are some things you should know regarding the hierarchy of storage locations within a computer and the principle of locality of reference. These facts will stand you in good stead no matter which language you choose to work in.
The Memory Hierarchy and Exploiting Locality of Reference
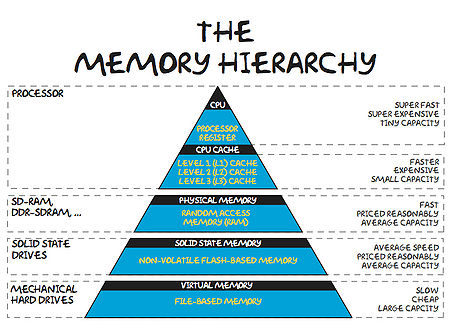
In order for it to operate, a computer must take data (from disk or memory), move it to the CPU, perform the relevant operations and have somewhere to store the results (again disk or memory). Now, it turns out that performing the operations takes far less time that retrieving the data from main memory. That being the case, computer designers have tried to level the playing field a little by adding layers of cache storage between the CPU and main memory. The caches are much quicker to access, but the trade-off is that they have relatively small capacities. The situation is summarised in the diagram and table below.
Access time analogy:
| L1 Cache | Picking up a book off your desk (~3s) |
| L2 Cache | Getting up and getting a book off a shelf (~15s) |
| Main Memory | Walking down the corridor to another room (several minutes) |
| Disk | Walking the coastline of Britain (about a year) |
The reason why caches ameliorate the situation is the observation that data access most often follow a pattern which is consistent with the principle of Locality of Reference.
We can identify two forms of locality:
- Temporal locality: We expect to re-use of data already seen.
- Spatial locality: We expect to access data stored close to data that we've already seen.
With this in mind, we will get the most from our hardware if we design our software accordingly.
Sort and Search
Hash tables (e.g. python dictionaries or sets) can provide very fast O(1) look up times, but come at the cost of some memory overhead (for python, perhaps more than 6x the size of list).
If you can't avoid the extra space and you'll need to store your data as a list (array). A linear lookup through an unordered list takes, of course, O(n) time for a list of length n. If you are going to perform more than one look up (and your data set is large), it will be (well) worth your while sorting the list first. Using e.g. the quicksort algorithm, the sort will take, on average, O(n log n) time, and using e.g. binary search, the look up will take, on average O(log n) time. Where 2(log n) << 2n, for large n.
There are many library routines, modules and packages which can provide you with sort and search facilities, so do have a poke about.
Not Exceeding RAM
One situation which gives poor performance is when a program's memory requirements exceeds the available RAM. If this happens, the operating system will attempt to keep the program running by swapping some of the data stored in RAM into a page file that is stored on disk. This operation comes are considerable cost to program performance and so should be avoided if at all possible.
Here is a little example program that can use up a lot of memory (depending upon the value of N), and has a random access pattern, so any part of the array stored in a page file on disk will be swapped back into RAM, sooner or later.
#include <stdio.h>
#include <stdlib.h>
#define N 500000000
int main()
{
int *bigArray;
int r;
bigArray = (int *)malloc(sizeof(int)*N);
while(1) {
r = rand()%N;
bigArray[r] = r;
}
return(EXIT_SUCCESS);
}
The program will keep running in an infinite loop, randomly accessing cells in a large array, until it is killed.
Once the program is running, the top command can give us a useful look at the machine resources and the state of any running processes:
Tasks: 170 total, 2 running, 168 sleeping, 0 stopped, 0 zombie Cpu(s): 51.3%us, 0.8%sy, 0.0%ni, 47.8%id, 0.0%wa, 0.0%hi, 0.0%si, 0.0%st Mem: 3341536k total, 3213400k used, 128136k free, 286092k buffers Swap: 0k total, 0k used, 0k free, 536384k cached PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND 8855 gethin 20 0 1908m 1.9g 272 R 100 58.5 0:47.21 foo.exe ...
In this case, we can see that the program foo.exe is showing good CPU utililisation (100%) and that the machine is not using any swap space.
The vmstat command, where si shows the amount of data swapped-in to RAM from disk, and so shows the amount swapped out of RAM onto disk:
gethin@gethin-desktop:~$ vmstat 1 procs -----------memory---------- ---swap-- -----io---- -system-- ----cpu---- r b swpd free buff cache si so bi bo in cs us sy id wa 1 0 0 98900 286340 500068 0 0 22 24 130 473 2 1 97 1 1 0 0 95420 286340 503984 0 0 0 0 657 758 51 1 49 0 1 0 0 95296 286340 504128 0 0 0 20 735 1198 50 0 49 0 1 0 0 95296 286340 503988 0 0 0 0 712 1142 51 0 49 0 1 0 0 95172 286340 504136 0 0 0 0 699 921 51 0 50 0 ...
Again, we see no swapping activity.
If we increase N, however, (to 2147483647); recompile and re-run, we see from top that the memory usage has, of course, increased and the %CPU has plummeted to just 1%!:
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND 320 root 10 -5 0 0 0 D 9.3 0.0 3:38.32 kswapd0 23033 ggdagw 18 0 8195m 6.7g 284 R 1.0 90.9 0:15.25 foo.exe ...
vmstat tells us a similar tale, where we see activity in the si and so collumns:
procs -----------memory---------- ---swap-- -----io---- --system-- -----cpu------ r b swpd free buff cache si so bi bo in cs us sy id wa st 0 3 518932 29272 1980 69412 0 0 25 3 0 0 6 0 94 0 0 1 3 518936 34320 1964 70212 2672 48 4128 48 465 506 0 1 87 12 0 0 2 518936 32348 1976 70700 2616 0 3800 36 596 848 0 0 87 13 0 1 3 518940 31052 1976 71964 2792 4 4324 4 482 533 0 0 88 12 0 1 4 518944 31292 1968 72028 2928 4 4404 4 539 569 0 1 87 12 0 0 4 518944 27848 1972 75332 2376 0 5804 0 558 806 0 0 87 12 0 0 2 523736 31356 1816 74292 3744 4796 4836 4960 636 579 0 3 87 10 0 0 3 523744 31788 1716 75256 2324 8 3768 56 461 493 0 1 87 12 0 ...
If you are working on bluecrsytal, you can check the memory & CPU usage of your program by first determining the node(s) that your job is running on, which qstat -n <jobid>, then logging into the node(s) and running the above top and vmstat commands.
Bespoke Applications
If you are using some 3rd party programs to process your data--perhaps supplied to you by a collaborating research group, do come and talk to the folks in the ACRC, as we may well be able to help you speed up your data processing.
Metadata, Data Curation and Data Re-Use
As the name suggests, metadata suggests, it is data about data. OK, fine. But why do we want it? Well, data comes at a price. There is the cost of creation (e.g. a trip to Antarctica to drill ice-cores) and there is a cost to storing it (e.g. buying servers & disks, paying the electricity bill and the salaries of system administrators). That being the case, we'd we wise to make the best use of it, once we have it.
When Data gets Big
- Quotas.
- seek vs read time
- seeks can add up, and dominate if files are small. To minimise:
- Make use of contiguous access patterns, rather than random ones.
- stream through large files
- Map-Reduce to process large volumes of data quickly.
- Hadoop is a popular, open-source implementation.
- Available on DICE. Please don't run on bluecrystal
Hadoop & Friends
Streaming MapReduce
mapper.py:
#!/usr/bin/env python
import sys
# input comes from STDIN (standard input)
for line in sys.stdin:
# remove leading and trailing whitespace
line = line.strip()
# split the line into words
words = line.split()
# increase counters
for word in words:
# write the results to STDOUT (standard output);
# what we output here will be the input for the
# Reduce step, i.e. the input for reducer.py
#
# tab-delimited; the trivial word count is 1
print '%s\t%s' % (word, 1)
reducer.py:
#!/usr/bin/env python
from operator import itemgetter
import sys
current_word = None
current_count = 0
word = None
# input comes from STDIN
for line in sys.stdin:
# remove leading and trailing whitespace
line = line.strip()
# parse the input we got from mapper.py
word, count = line.split('\t', 1)
# convert count (currently a string) to int
try:
count = int(count)
except ValueError:
# count was not a number, so silently
# ignore/discard this line
continue
# this IF-switch only works because Hadoop sorts map output
# by key (here: word) before it is passed to the reducer
if current_word == word:
current_count += count
else:
if current_word:
# write result to STDOUT
print '%s\t%s' % (current_word, current_count)
current_count = count
current_word = word
# do not forget to output the last word if needed!
if current_word == word:
print '%s\t%s' % (current_word, current_count)
Place some text files into input dir in HDFS.
Script to run it:
#!/bin/bash export HADOOP_MAPRED_HOME='/opt/cloudera/parcels/CDH-4.3.0-1.cdh4.3.0.p0.22/lib/hadoop-0.20-mapreduce' hadoop jar $HADOOP_MAPRED_HOME/contrib/streaming/hadoop-streaming-2.0.0-mr1-cdh4.3.0.jar \ -file /home/ggdagw/mapper.py \ -mapper mapper.py \ -file /home/ggdagw/reducer.py \ -reducer reducer.py \ -input /user/ggdagw/input/* \ -output /user/ggdagw/output-wc-streaming
Hive
Hive is an SQL-like interface to data stored in HDFS. Below is an example of using Hive with the planets table that we saw earlier when considering SQLite and MySQL.
[ggdagw@hd-37-00 ~]$ hive hive>
hive> CREATE DATABASE menagerie;
OK
Time taken: 0.115 seconds
hive> use menagerie;
OK
Time taken: 0.061 secondshive> CREATE TABLE planets (Id INT, Name STRING,
> Diameter FLOAT,
> Mass FLOAT,
> Orbital_Period FLOAT)
> ROW FORMAT delimited fields terminated by ',' STORED AS TEXTFILE;
hive> LOAD DATA LOCAL INPATH 'planets.csv' INTO TABLE planets;
hive> SELECT * FROM planets;
OK
1 'Mercury' 0.382 0.06 0.24
2 'Venus' 0.949 0.82 0.72
3 'Earth' 1.0 1.0 1.0
4 'Mars' 0.532 0.11 1.52
5 'Jupiter' 11.209 317.8 5.2
6 'Saturn' 9.449 95.2 9.54
7 'Uranus' 4.007 14.6 19.22
8 'Neptune' 3.883 17.2 30.06
Time taken: 0.281 secondsSummary
- Use large files whenever possible.
- Disks are poor at servicing a large number of seek requests.
- Check that you're making best use of a computer's memory hierarchy, i.e.:
- Think about locality of reference.
- Go to main memory as infrequently as possible.
- Go to disk as infrequently as possible as possible.
- Check that your are still using the right tools if your data grows.